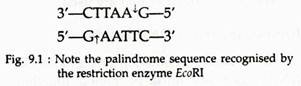In this article we will discuss about:- 1. Meaning of Recombinant DNA Technology 2. Basic Steps in Recombinant DNA Technology 3. Preparation.
Meaning of Recombinant DNA Technology:
In nature, exchange of information between two genetic materials resulting in acquisition of new segments of DNA by a genome is very common, but it occurs only among genetic materials of the same or closely related species.
In the last several decades, thousands of scientists working in genetics, biochemistry, cell biology and physical chemistry have yielded techniques to construct recombinant molecules containing foreign DNA into cells of unrelated organism. A specific characteristic of one organism would then be visible in a different organism resulting in a living chimera. Thus things have happened that were not even dreamt of.
The deliberate modification of an organism’s genetic information by directly changing its nucleic acid genome is called genetic engineering. Genetic engineering is accomplished by a collection of methods known as recombinant DNA technology. First the DNA responsible for a particular phenotype is identified and isolated.
ADVERTISEMENTS:
Once purified, the gene or genes are fused with other pieces of DNA to form recombinant DNA molecules or rDNA. The term recombinant DNA usually refers to the creation of new combination of DNA molecules that are not found together naturally.
Although processes such as crossing over technically produce recombinant DNA, the term is generally reserved for DNA molecules produced by joining molecules derived from different biological sources.These rDNAs are then propagated or cloned by insertion into an organism that need not even be in the same kingdom as the original gene donor.
Techniques for DNA cloning — either by cell- based method or cell-free in vitro method — have opened hitherto unimaginable opportunity to identify and study the genes involved in almost every known biological process.
Recombinant DNA technology or genetic engineering has opened up totally new areas of research and applied biology. Thus, it is an essential part of biotechnology which is now experiencing a stage of exceptionally rapid development and progress.
Basic Steps in Recombinant DNA Technology:
ADVERTISEMENTS:
Recombinant DNA technology is a powerful tool for the isolation of potentially unlimited quantities of a gene.
The basic procedure involves the following steps:
(i) Purification of DNA from cells or tissues or construction of cDNA (complementary DNA) from total RNA or poly(A)+ messenger RNA (mRNA) using the enzyme reverse transcriptase.
(ii) Production of DNA fragments (for genomic DNA, not for cDNA) using the enzymes called restriction endonucleases that recognise and cut DNA molecules at specific nucleotide sequences.
ADVERTISEMENTS:
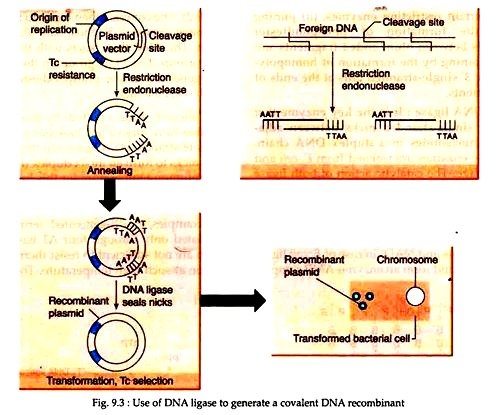
(iii) Joining of DNA fragments or cDNA using the enzyme ligase to other DNA molecules that serve as vectors or carrier molecules, having capacity of independent replication. For joining genomic DNA fragments, cutting the target DNA and the vector will be done by the same restriction enzyme. The vector with an inserted DNA segment is a recombinant DNA molecule.
(iv) Transformation of the recombinant DNA molecule, carrying an inserted DNA fragment into a host cell (often bacterial or yeast cells) in which the chosen molecule can undergo replication, independently of the host cell chromosomes and produce dozens of identical cells known as clone.
As host cells replicate, the recombinant DNA molecules within them are passed on to all their progeny cells, creating a population of host cells, each of which carries copies of a cloned DNA sequence.
(v) Isolation of the recombinant DNA clones by harvesting the expanded cell cultures; subsequent purification and analysis of that target DNA. Potentially, the cloned DNA can be transcribed, its mRNA translated and the gene product isolated and used for research or sold commercially.
Preparation of Recombinant DNA Technology:
Before describing the methods of preparation of recombinant DNA molecules, concepts about three key elements viz, restriction endonuclease, DNA ligase and vectors that are essential for recombinant DNA technology are given below:
Restriction Enzymes:
The corner stone of genetic engineering or recombinant DNA technology is a special class of enzymes called restriction enzymes. These are bacterial enzymes that recognise specific nucleotide sequences, called restriction sites, and cut both strands of DNA at this site.
Since these enzymes cleave DNA within the molecule, they are called restriction endonucleases to distinguish them from exonuclease enzymes which digest or cleave nucleic acids from an end.
These enzymes got the name restriction because these enzymes naturally occur in bacteria where they prevent or restrict viral infection by degrading the invading viral DNA. The term restriction endonuclease was coined by Lederberg and Meselson (1964). In 1978, Werner Arber, Hamilton Smith and Daniel Nathans received Nobel Prize for their work on these enzymes.
ADVERTISEMENTS:
To date, about 350 restriction enzymes have been identified and purified from several hundred different species of bacteria. In genetic engineering technology, restriction endonucleases are popularly called as molecular knives or molecular scissors or molecular scalpels due to their ability to reproducibly cut DNA into fragments.
Naming of restriction endonucleases According to Smith and Nathans (1973) naming of the restriction enzymes is based on following rules:
(a) Each enzyme-name should have a three-letter code.
(b) The first letter of the code name is derived from the first epithet of the genus name of the bacteria from which it has been obtained.
(c) The second and third letters are from the species name of the bacteria.
(d) The three-letter code name is followed by the strain number. If one strain has more than one restriction enzymes, these will be identified by Roman numerals as I, II, III etc.
(e) A general name endonuclease R may be added, e.g., EcoRI enzyme which is derived from E. coli carrying antibiotic resistance plasmid (RI).
Types of restriction enzymes:
I. Type I restriction enzymes:
Some restriction enzymes interact with an unmodified recognition sequence in double strand DNA and then attach to long DNA molecule. After travelling far distance between 1,000 to 5,000 nucleotides, the enzyme cleaves only one strand of the DNA at an apparently random site, and creates a gap of about 75 nucleotides in length.
Since, the site of cleavage of these enzymes are not specific, these enzymes are not useful for gene manipulation. The type I enzymes can hold two different sites on DNA, simultaneously creating a loop in nucleic acid. These enzymes consist of three subunits, e.g., the EcoK has R2M2S sub- units. The R subunit is responsible for restriction, the M subunit for methylation and the S subunit helps in binding of enzyme to DNA.
2. Type II restriction enzymes:
Unlike type I enzymes these restriction enzymes recognise a particular sequence in the double- stranded DNA molecule and cleave the chain within or near that sequence to give rise to discrete DNA fragments of defined length and sequence. These enzymes require Mg2+ ions for their function e.g., EcoRI.
3. Type III restriction enzymes:
These enzymes possess intermediate properties between type I and type II enzymes. They cleave double stranded DNA at specific sites. They require ATP, Mg2+ and partially S- adenosylmethionine for their activities e.g., EcoPI, Eco15 etc.
Mechanism of action of restriction enzymes:
1. Target sites:
Restriction enzymes of type II recognise a specific base sequence of four to six nucleotides long and make a cut at that site only. Most of these recognition sites are palindrome sequence within the DNA duplex. Palindrome sequences are sequences that read the same in both directions (see Fig. 9.1).
This segment is said to have two fold rotational symmetry because it can be rotated 180° without change in base sequence. When an enzyme attacks this palindrome, it breaks each strand at the same site in the sequence, which is indicated by the arrows between the A and G residues as shown:
2. Nature of the cut-ends:
The restriction enzyme cuts DNA molecules by cleavage in two fashions:
(a) Blunt-end fashion, and
(b) Sticky- or cohesive-end fashion.
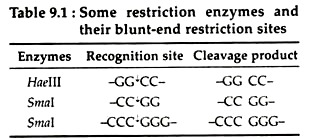
(a) Blunt-end fashion:
Certain restriction enzymes like Smal, Haelll cleave DNA duplex at the same position to produce blunt end fragments. The utility of this style during the joining of DNA fragments is that any pair of ends may join together, irrespective of sequence. This is useful to join two defined sequences without introducing any additional material between them (Table 9.1):
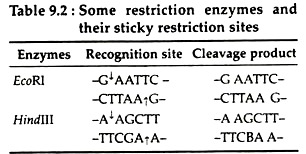
(b) Sticky- or cohesive-end fashion:
Most restriction enzymes like EcoRI, BamHl, Hindlll make single strand staggered cut at some nucleotide pairs apart in the opposite strands of DNA and generate fragments having “Single stranded trail’ or ‘Sticky- ends’ at both ends.
These fragments generated at a given restriction site are complementary to those on all other fragments generated by the same enzyme. At room temperature, these single-stranded fragments can transiently base pair with those of other DNA fragments generated with the same restriction enzyme, regardless of the source of the DNA (Table 9.2):
3. Restriction fragment:
Restriction enzymes cut the DNA into a reproducible set of fragments called restriction fragments. Usually restriction enzymes recognise a specific base sequence on the DNA and make a cut at this site only; these base sequences are called restriction recognition sites. These are short DNA sequences having four to eight base pairs.
4. Star activity:
Some restriction endonucleases may show relaxation in specificity of sequence under non-optional conditions. During such condition, one enzyme even can recognise other alternative base instead of its specific base. Non ideal ionic strength in buffers, high glycerol concentration and high enzyme concentration has been shown to alter the DNA recognition sequence for many restriction enzymes.
Modification enzymes, Isoschizomers and Host-controlled Restriction:
The restriction sites in the chromosome of a bacterium are protected from its own restriction endonuclease by a special type of enzyme called modification enzyme. This enzyme adds a methyl group to one or two bases, usually within the restriction site. It methylates adenine in the N-6 position and cytosine either in the N-5 or N-4 position and produce N-6 methyl adenine and 5 or 4- methyl cytosine, respectively.
If a methyl group is present there, the restriction enzyme is prevented from cutting the DNA. Together with the restriction endonuclease, the methylene enzyme (methyltransferase) forms a restriction-modification system; that protects the host DNA while it destroys foreign DNA.
Isoschizomers are restriction enzymes that are isolated from different organisms but recognise identical base sequences in the DNA. For example, Asp718, isolated from Achromobacter species 718 and Kpnl, isolated from Klebsiella pneumoniae OK8 have identical recognition sites.
Certain strains of bacteria are immune to bacteriophages. This is called host-controlled restriction. This restriction is due to some endonucleases which could recognise and split specific loci in the foreign DNA. Thus, these enzymes prevent or restrict the survival of foreign DNA in the host.
Importance of Restriction enzymes in Genetic Engineering:
Restriction enzymes are the basic tools of genetic engineering.
Their importance in genetic engineering are:
(i) The use of different restriction enzymes allows the DNA of human genome, or that of other organisms to be dissected into a precisely defined set of specific fragments.
(ii) The number and size of the fragments generated by a restriction enzyme tells the frequency of occurrence of the target site in that DNA.
(iii) In order to join two fragments of DNA together, it is not essential that they are produced by the same restriction enzymes. Many different restriction enzymes produce compatible cohesive ends. For example, Age I and Aval produce molecules with identical 5′ overhangs and so can be ligated together.
(iv) Methylation can reduce the susceptibility of DNA to cleavage by restriction endonucleases and the efficiency of DNA transformation.
(v) It is important to eliminate restriction systems in E. coli strains used as hosts for recombinant DNA. If the foreign DNA is introduced into an E. coli host it may be attacked by restriction systems active in the host cell. Some strain like E. coli C and HB 101 have deletion in restriction system and hence lack all restriction activities.
(vi) The success of a cloning experiment is critically dependent on the quality of the restriction enzymes that are used.
Joining enzymes:
DNA Ligase:
To create a recombinant DNA molecule, DNA fragments must be joined together.
There are currently three methods for joining the DNA fragments in vitro:
(i) Joining covalently the annealed cohesive ends produced by certain restriction enzymes,
(ii) Joining by the formation of phosphodiester bonds between blunt ended fragments and
(iii) Joining by the formation of homo-polymeric 3′ single-stranded tails at the ends of fragments.
DNA ligase:
It is the key enzyme that seals single-stranded nicks between adjacent nucleotides in a duplex DNA chain. Ligase enzymes are isolated from E. coli and T4 phage. The catalytic action of both ligase enzymes is very similar, but they differ in their cofactor requirements. T4 ligase requires ATP, while E. coli ligase requires NAD+.
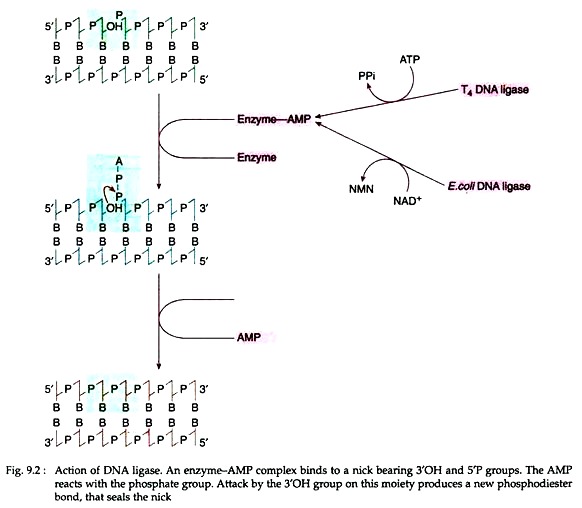
Mechanism of action:
ATP in case of T4 DNA ligase and NAD+ in case of E. coli ligase first split and form an enzyme-AMP complex (Fig. 9.2). The complex then binds to the nick that must expose a 5′ phosphate and 3’OH group. The AMP then reacts with the phosphate group. The 3’OH group on the moiety then generates a new phosphodiester bond which seals the nick.
Cohesive ends created by restriction enzymes have nicks a few base pairs apart in opposite strands. DNA ligase can then repair these nicks to form an intact duplex (Fig. 9.3). The optimum temperature for this ligation is 37°C, but at this temperature, the hydrogen- bonded between the sticky ends is unstable.
For examples, EcoRI-generated termini get associated only through four AT base pairs which are not sufficient to resist thermal disruption at such high temperature. Therefore, the ligating temperature is a compromise between the rate of enzyme action and association of the termini. It has been found to be in the range of 4-15°C.
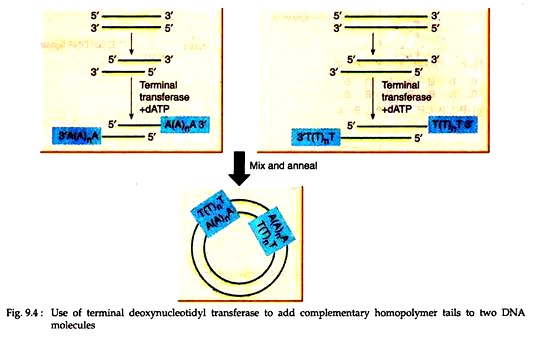
Joining of blunt ended DNA molecules is mainly done by annealing complementary homopolymer sequences. Thus, by adding oligo (dA) sequences to the 3′ ends of one population of DNA molecules and oligo (dT) blocks to the 3′ ends of another population, the two types of molecules can be annealed to form mixed dimeric circles (Fig. 9.4).
The enzyme terminal deoxynucleotidyl transferase, purified from calf thymus, provides the means by which the homo-polymeric extensions can be synthesised. It can repeatedly add nucleotides to the 3’OH termini of a DNA molecule.
DNA with exposed 3’OH can be generated by pretreatment with phage exonuclease or restriction enzyme Pestle. In recent years, homopolymer tailing has been largely replaced as a result of the availability of a much wider range of restriction endonuclease and other DNA modifying enzymes. However, it is still important for cDNA cloning.
Prevention of recircularisation:
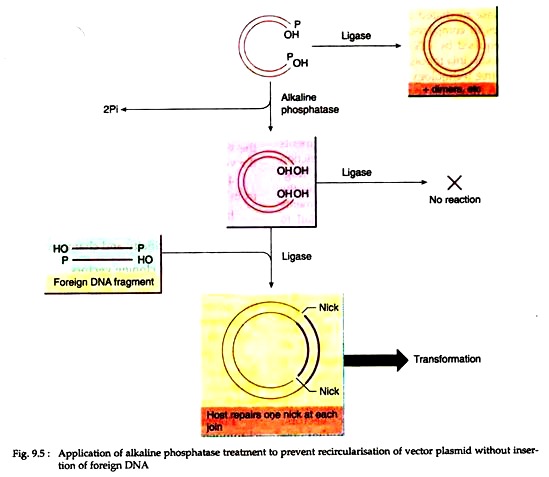
During cloning, the vector DNA molecule following cutting should be treated with alkaline phosphatase. It will remove 5′ terminal phosphate groups, thus preventing recircularisation and plasmid dimer formation (Fig. 9.5).
In this case, circularization of the vector can occur only by insertion of non-phosphatase treated foreign DNA which has one 5′ terminal phosphate at each join. One nick at each join remains un-ligated, but after transformation of host bacteria, cellular repair mechanisms reconstitute the intact duplex.
Joining DNA molecules without ligase:
The enzyme vaccinia topoisomerase can alone cause cleavage and joining of DNA molecules. When CCCTT cleavage motif is placed near the end of a duplex DNA, the enzyme generates a stable, highly recombinogenic protein DNA adduct that can only religate to acceptor DNAs, that contain complementary single-strand extensions.
Thus, linear DNAs containing CCCTT cleavage sites at both ends can be activated by topoisomerase and inserted into a plasmid vector. Topoisomerase mediated ligation requires only 5 minutes in comparison to overnight incubation required by DNA ligase. For these properties vaccinia topoisomerase can be used for ligase-free technology for the covalent joining of DNA fragments to plasmid vectors.
Vectors:
Cutting of DNA molecules into fragments is possible with the help of specific restriction endonuclease enzymes. These DNA fragments can be readily joined together by the activities of DNA ligase enzymes. If fragments of DNA are inserted into cells, they fail to replicate except in those rare cases where they integrate into the chromosome. To enable such fragments to be propagated, they are inserted into DNA molecules (vectors) that are capable of extra-chromosomal replication.
Originally, the purpose of vectors was the propagation ‘of cloned DNA but today vectors fulfill many other roles, such as facilitating DNA sequencing, promoting expression of cloned genes, facilitating purification of cloned gene products and reporting the activity and localisation of proteins. There are two basic vectors, cloning and expression vectors that are most widely required for recombinant DNA technology.
Cloning Vectors:
These are autonomously replicating genetic elements used to carry a cDNA or fragment of genomic DNA into a host cell for the purpose of gene cloning. DNA fragments produced by restriction enzyme digestion cannot directly enter the bacterial cells, but when a DNA fragment is joined to a vector, however, it can gain entry to a host cell. Vectors are, therefore, carrier of DNA molecules.
However, to serve as a vector, a DNA molecule must possess the following properties:
1. Vector must be able to independently replicate itself and the carried DNA segment.
2. Vector should contain several restriction enzyme cleavage sites that are present only once in the vector. The restriction enzyme that cleaves this site should also be used to cut the DNA fragment to be inserted into the vector. That means the same restriction enzyme/ enzymes will be used to cleave both vector as well as DNA fragments to be inserted.
3. Vector should carry suitable marker (usually in the form of antibiotic resistance genes or genes for enzymes missing in the host cell) to distinguish host cells that carry the vectors from host cells that do not carry the vectors.
4. It should be easy to obtain vectors from their host cells.
In recent years many vectors are being used in recombinant DNA technology, each with its own specificity and characteristics.