In this article we will discuss about DNA:- 1. The Physical Structure of DNA 2. Properties of DNA.
The Physical Structure of DNA:
The Double Helix:
From 1940 to 1953, many scientists were interested in unraveling the structure of DNA molecule. They were Erwin Chargaff, Maurice Wilkins, Rosalind Franklin, Linus Pauling, Francis Crick and James Watson. In 1953, two of them James Watson and Francis Crick, proposed that the structure of DNA is in the form of a double helix.
Their proposal was published in a short paper in the journal “Nature”. The data available to Watson and Crick came primarily from two sources — base composition analysis of hydrolysed samples of DNA and X-ray diffraction studies of DNA.
ADVERTISEMENTS:
Their proposed model is based on the following major features:
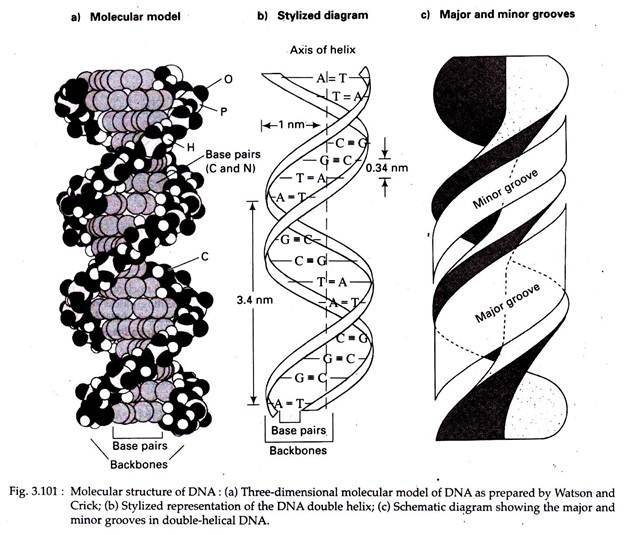
1. The DNA molecule consists of two long polynucleotide chains coiled around each other in a right-handed fashion (alternative form of DNA may also exist).
2. The two chains are antiparallel, that is, their C-5′ to C-3′ orientations run in opposite directions.
3. The diameter of the helix is 2 nm.
ADVERTISEMENTS:
4. The sugar-phosphate backbones are on the outside of the double helix, while the bases are oriented towards the central axis (Fig. 3.101b). The bases of both chains are flat structures, lying perpendicular to the long axis of the DNA; that is, the bases are ‘stacked’ on one another, except for the ‘twist’ of the helix (Fig. 3.101a).
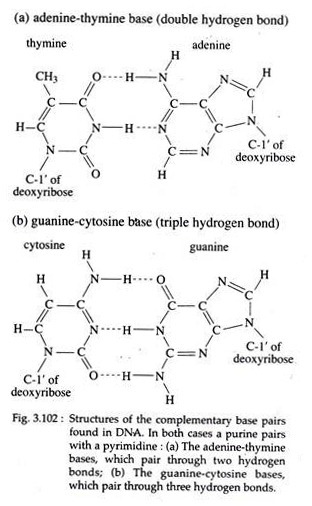
5. The nitrogenous bases of opposite chains are bonded together by relatively weak hydrogen bonds in DNA. The specific pairing observed are A with T by two hydrogen bonds and G with C by three hydrogen bonds. These specific A-T and G-C pairs are called complementary base pairs (Fig. 3.102).
6. The base pairs are 0.34 nm apart from each other in the DNA helix. A complete turn (360°) of the helix is 3.4 nm; thus there are 10 bases per turn. Each base pair, then, is twisted 36° clockwise with respect to the previous base pair.
ADVERTISEMENTS:
7. Because of the way the bases bond with each other, the two sugar phosphate backbones of the double helix are not equally spaced along the helical axis. This results in- grooves of unequal size between the backbones called major groove (the wider groove of the two) and minor groove (the narrower groove of the two) (Fig. 3.101c).
The nature of base pairing is the most important feature of the model. Why aren’t other base pairs possible? Watson and Crick discounted the A=G or C=T pairing, because these represent purine—purine and pyrimidine—pyrimidine pairings, respectively.
Such pairing would lead to alternating diameters of more than and less than 2 nm because of the respective sizes of the purine and pyrimidine rings. Moreover, the three dimensional configuration formed by such pairing do not produce the proper alignment leading to sufficient hydrogen bond formation.
For this reason A=C and G=T pairing were also bases, which pair through two hydrogen bonds; (b) The guanine-cytosine bases, which pair through three hydrogen bonds discounted, even though in these pairs each consists of one purine and one pyrimidine.
A more recent and accurate analysis of the form of DNA revealed a minor structural difference. The number of base pairs per turn has demonstrated a value of 10.4 rather than the 10.0 predicted by Watson and Crick because the new finding requires a rotation of 34.6° for each complete turn instead of 36°.
However, the ideas proposed by Watson and Crick on DNA structure were a remarkable feat and were highly significant to subsequent studies in genetics and biology. Recognition of the work leading to the double-helical model subsequently came in the form of Nobel Prize, awarded in 1962 to Watson, Crick and Wilkins.
Other DNA Structures:
A more thorough analysis by single- crystal X-ray diffraction of the early X-ray diffraction patterns of DNA fibres reveals that DNA can also exist in different form, such as:
ADVERTISEMENTS:
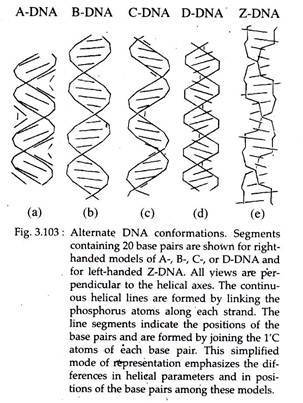
(i) A-DNA and B-DNA:
A-DNA and B- DNA are right-handed double helices with 10.9 and 10.0 base pairs per 360° turn of the helix, respectively. The bases are tilted and displaced laterally in relation to the axis of the helix (which would be 0°) by 13° in A-DNA and 2° in B-DNA. A-DNA double helix is short and wide with a narrow, very deep major groove and a wide, shallow minor groove (Fig. 3.103a).
The B-DNA double helix is thinner and taller for the same number of base pairs than A-DNA, with a wide major groove and a narrow minor groove; both grooves are of similar depth (Fig. 3.103b). B-DNA is the form typically found in cells (as described by Watson and Crick) and A-DNA is found only when the DNA is in dehydrated states.
Three other right-handed forms of DNA helices have been discovered and designated as C-, D- (Fig. 3.103), and E-DNA. C-DNA has only 9.3 base pairs per turn and thus is less compact. Its base pairs are tilted relative to the axis of the helix. D-DNA and E-DNA lack guanine in their base composition and they have 8.0 and 7.5 base pairs per turn, respectively.
(ii) Z-DNA:
Z-DNA is a left-handed double helix (Fig. 3.103e). There are 12.0 base pairs per complete helical turn. The bases are inclined perpendicularly away from the helix axis by 8.8 degrees. The Z-DNA helix is thin and elongated with a deep, minor groove.
The major grooves are pushed to the surface of the helix making them indistinct. Indirect evidence for the presence of Z-DNA has come from the identification of Z-DNA-binding proteins in cells, e.g., in nuclei of Drosophila, human, wheat and bacterial cells. It is proposed that Z-DNA has a role in at least some replication and transcription events and in local DNA unwinding, required for genetic recombination events.
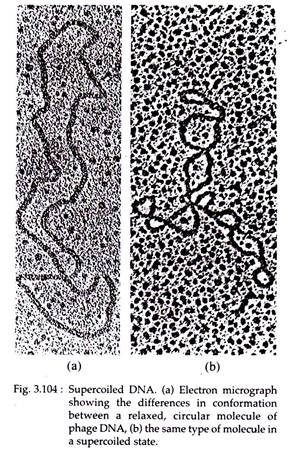
Supercoiled DNA:
In 1963, Jerome Vinograd found that circular DNA of bacteria and viruses can exist in a relaxed or a super- coiled form (Fig. 3.104). The supercoiled DNA may be of two types — positive, when helix is in right handed coil and negative, when the double helix is in left-handed coil.
Properties of DNA:
1. Absorbance:
The nitrogenous bases of the nucleic acids absorb UV radiation most strongly at wavelengths of 254 to 260 nm due to interaction between UV light and the ring systems of the purines and pyrimidine’s. Actually the different nucleotides absorb UV of following wavelengths: A-262.5 nm, T-264.5 nm, G-249 nm and C-276 nm.
The absorbance of DNA is about 1.0, but that of RNA and single-stranded DNA is about 1.2-1.3. When DNA is denatured, its absorbance power increases than in native state. This is called hyper-chromic shift.
2. Denaturation and Renaturation:
The double helical structure of DNA is preserved by weak interactions; so it is possible to separate the two strands by treatments involving heating or alkaline pH. This separation is called melting or denaturation of DNA.
The temperature at which DNA strands separate (the melting point) depends on the AT/GC ratio. If the DNA is cooled slowly after denaturation, the complementary strands will base pair and the native (double helical) conformation will be restored.
This process is called renaturation or annealing. Renaturation of DNA can be used to estimate the size (number of nucleotides) of the genome of a given organism. Renaturation studies led to the discovery of repeated sequence in eukaryotic DNA; the presence of which causes the rate of renaturation much faster than for sequences present as single copies.
On the basis of the kinetics of renaturation, eukaryotic DNA falls into three categories like highly repeated DNA sequences, moderately repeated DNA sequences and non-repeated DNA sequences. The first group of DNA sequences re-anneal very rapidly and are very short in length. These include following types:
(i) Satellite DNAs:
These DNAs consist of short sequences (from about 5 to 100 base pairs in length) that are repeated many times to form very large clusters containing up to 100 million base pairs of DNA.
(ii) Mini-satellite DNAs:
These DNAs average about 15 base pairs in length and are found in clusters containing as many as 1,000 to 3,000 repeats.
(iii) Microsatellite DNAs:
These are short sequences (2 to 5 base pairs long) and are present in clusters of 100 or fewer repeated copies. Moderately repeated DNA sequences are repeated within the genome anywhere from a few times to several hundred thousand times. Included among these DNA sequences are ones that code for known gene products, either RNAs or proteins, and those that lack a coding function.
Non-repeated DNA sequences (70 percent of human DNA fragments of 1,000 base-pair length) are very slow to find partners and are presumed to be present in a single copy per genome. They are present in a particular site on a particular chromosome. Single-stranded DNA can also anneal to complementary RNA, resulting in a hybrid molecule.
Such molecular hybridization is a very powerful technique for characterizing RNAs. Denaturation of DNA, on the other hand, may be used for physical mapping of DNA in a technique called partial denaturation mapping. The regions rich in AT separate more easily. Under EM, these regions are detected as single-stranded loops, and the distance between the loops and the end of the DNA’ molecule can be measured.
3. Sedimentation Behaviour:
DNAs of different densities can be separated by subjecting them to density gradient centrifugation. The technique can also be used to generate data on the base composition of double stranded DNA. The percentage of A=T or G≡C pairs in DNA is directly proportional to the molecule’s buoyant density.
Molecular characterization of DNAs from different sources can be done by this useful technique. Another technique, sedimentation velocity centrifugation can be used to determine the ‘velocity of sedimentation’ of DNA molecules that in turn is used to determine the molecular weight.
4. Solubility:
DNA is dissolved in water and alcohol. In chilled alcohol it is insoluble and thus become visible.
5. Fuelgen Reaction:
DNA if treated with warm acid (hydrolysis), a reddish- purple staining reaction would occur upon the addition of Schiff’s reagent. This reaction is called Fuelgen reaction after the name of the discoverer and it has been found to be specific only for DNA.
Neither RNA nor any other cell substance will give the precise DNA type of staining reaction. 6. Ionic property being negatively charged DNA moves towards anode in the electric field. On the basis of this ionic property, DNA can be separated by gel electrophoresis.