In this article we will discuss about RNA:- 1. Structure of RNA 2. Major Classes of RNAs 3. Some Special RNAs 4. Properties.
Structure of RNA:
The primary structure of RNA is similar to that of DNA, except for the substitution of ribose sugar for deoxyribose and base uracil for thymidine. RNA molecules consist of only one chain, so the base composition of RNA does not follow Chargaff’s rule (amount of adenine equals the amount of thymine, A=T and the amount of cytosine equals the amount of guanine, C=G).
There are three major classes of RNA all of which are involved in protein synthesis:
i. Messenger RNA (mRNA):
ADVERTISEMENTS:
Messenger RNA (mRNA), carries the genetic information for the sequence of amino acids. They vary considerably in size, which is reflected in the variation of the size of the protein encoded by mRNA as well as the gene serving as the template for transcription of mRNA.
ii. Transfer RNA (tRNA):
Transfer RNA (tRNA), is the smallest class of RNA molecules. It identifies and transports amino acid molecules to the ribosome during translation.
iii. Ribosomal RNA (rRNA):
ADVERTISEMENTS:
Ribosomal RNA (rRNA) represents 50% of the mass of ribosomes, that provide a molecular scaffold for the chemical reactions of polypeptide assembly. Although each RNA molecule has only a single polynucleotide chain, the structure of RNA is not simple, smooth or linear.
All RNA molecules have extensive regions of complementarity in which hydrogen bond between AU and GC pairs are formed between different regions of the same molecule. Structures formed as a result of folding of the molecule upon itself are called hairpin loops.
In this base-paired region the RNA molecule adopts a helical structure comparable to that
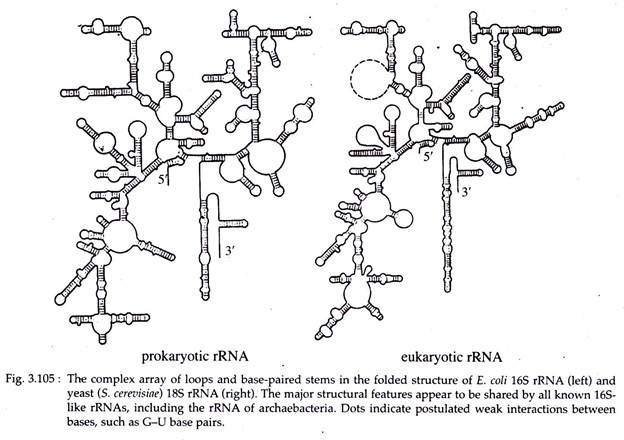
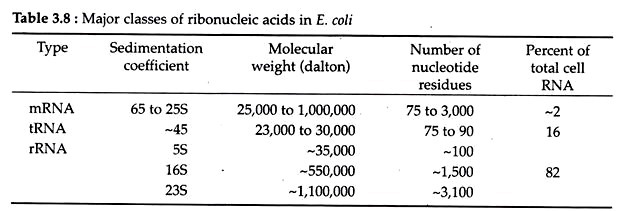
of DNA. The size, sedimentation behaviour (Svedberg co-efficient: S) of different RNA molecules are tabulated in Table 3.8.
 Major Classes of RNAs:
Major Classes of RNAs:
i. mRNA:
ADVERTISEMENTS:
An mRNA molecule after maturation consists of
(a) Coding region:
This is the main region with codons for amino acids. This region begins with 5′-AUG-3′ initiation codon and ends in 5′-UAA-3′ or UAG or UGA termination codon.
(b) Leader, cap and Shine-Dalgarno Sequence:
In between the initiation codon and extreme 5′ end, there is a non-coding sequence called leader sequence. It is a transcriptional product of the anti-leader sequence of the DNA template. The 5′ end of eukaryotic mRNA bears guanine nucleotide (most commonly 7-methyl guanosine or m7G) which is called the Cap.
In eukaryotes, three types of cap exist — Cap 0 (unicellular eukaryote), Cap 1 and Cap 2 (all eukaryotes except unicellular organisms). Cap is essential for binding of mRNA to the small subunit of ribosome during protein synthesis.
In case of prokaryotes and phages, the leader of mRNA bears a so-called ribosome binding sequence or Shine-Dalgarno sequence. The terminal region of 16S rRNA of small 30S ribosomal subunit of prokaryote at its 3′ end bear the sequence like – 3’…UCC UCC…5′.
The leader of mRNA in prokaryotes also contains a sequence which is partly or altogether complementary to this, and this is 5’…AGG AGG…3′. This poly-purine stretch has been called Shine-Dalgarno sequence. This sequence pairs by complementary base pairing with the 16S rRNA of 30S ribosomal small subunit during mRNA- ribosome binding at the onset of protein synthesis.
(c) Trailer sequence and poly-A tail:
ADVERTISEMENTS:
The coding region of mRNA ends at the terminator Condon which is either…UAA-3′ or …UGA-3′ or …UAG-3′. But the mRNA does not terminate there. Additional sequences that lie in between the termination codon and the extreme 3′ end is called the trailer sequence.
The terminal part towards 3′ end often consists of poly-A sequence which is not a transcriptional product but is added later. This poly-A sequence is found mostly in eukaryotes and called poly-A tail. It is suggested that the poly-A tail stabilizes the structure of mRNAs with which it is attached.
ii. rRNAs:
Eukaryotic cells contain millions of ribosomes, each of which contains several molecule of rRNA together with dozens of ribosomal protein. In prokaryotes, three type of rRNAs are available viz., 16S rRNA, 23S rRNA and 5S rRNA. The small ribosomal subunit contains 16S rRNA and the large subunit contains rest two types of rRNAs.
In eukaryotes, the large 60S subunit of ribosome contains 28S, 5S and 5.8S rRNA and the small 40S subunit contain 18S rRNA. Primarily rRNA is single stranded but 70% of its structure are secondarily double helical (Fig. 3.105).
iii. tRNA:
Among different types of RNAs, tRNAs are the best characterized RNA molecules. They are composed of only 75 to 90 nucleotides and display a nearly identical structure in bacteria to eukaryotes.
In 1965, Robert Holley and his colleagues reported the complete sequence of yeast alanine tRNA which included many unique nucleotides that are the modified bases expected in RNA (G, C, A and U). These include inosinic acid (containing purine hypoxanthine), ribo-thymidylic acid, pseudo- uridine (Ψ), methyl guanine, methylamine purine etc.
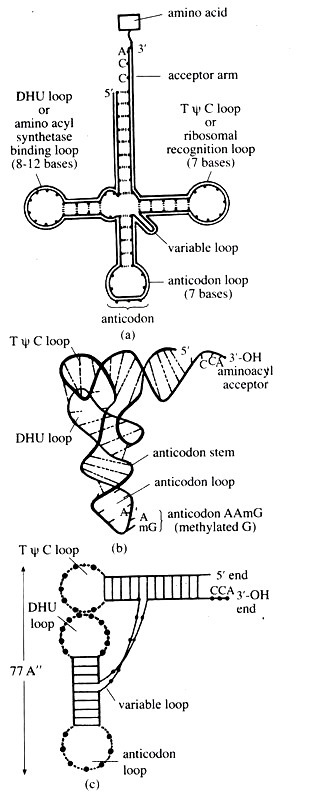
Holley argued and established that he could arrange the linear sequences of tRNA nucleotides in such a way that several stretches of base pairing would result in the formation of paired stems and unpaired loops resembling the shape of a clover-leaf. Holley’s clover-leaf model of tRNA is shown in Fig. 3.106a.
All tRNA molecules have the same terminal sequence pCpCpA – at the 3′ end where the amino acid is covalently joined to the terminal adenosine residue. All tRNAs contain 5′-Gp at the other end of the molecule.
The double helical stems with loop include:
(i) The acceptor arm that carries a single stranded region representing the 3′ acceptor end of tRNA with the base sequence 5’CCA 3′,
(ii) The TOC arm (or T arm) or ribosomal recognition arm with triplet sequence TWC in the loop carried in this arm,
(iii) D arm characterized by the presence of dihydrouridine (DHU) in the associated loop, and (iv) the anticodon arm that contains a specific triplet of nucleotide (anticodon) that recognizes its corresponding codon in mRNA.
The anticodon is different in the tRNAs for different amino acids. One of the base in the arrticodon is the wobble base which is less specific in its interaction with its corresponding base in the codon than the other two anticodon bases.
There may be also a highly variable region, called variable loop, which differs greatly in the different tRNAs. This extra loop lies between the WC and anticodon arms. On the basis of the size of the variable loop, tRNAs have been divided into two classes, (i) Class I tRNAs have a small variable loop with only 3 to 5 bases, (ii) Class II tRNAs have a large variable loop of 13 to 21 bases.
By 1974, different workers proposed the three dimensional structure (TDS) of tRNA by X-ray crystallographic examination. The TDS revealed an ‘If shaped structure with two major domains: (i) acceptor arm-TWC minihelix and (ii) anticodon arm-DHU (or D) arm-biloop (Fig. 3.106b, c).
Some Special RNAs:
i. hn RNA :
James Darnell and his coworkers first observed that eukaryotic mRNA is transcribed initially as a precursor molecule much larger than that which is translated. These RNAs are called pre-mRNAs or heterogeneous nuclear RNA (hn RNA).
These are of variable but large sized (up to 107 dal- tons) and are complexed with proteins, forming heterogeneous nuclear ribonucleoprotein particles (hnRNPs). About 25 per cent of hnRNAs are converted into mRNA, through complicated splicing processing.
ii. Sn RNA:
These small nuclear RNAs possess usually 100 to 200 nucleotides or less and are often complexed with proteins to form small nuclear ribonucleoproteins (SnRNPs or snurps). They are rich is uridine residues and are arbitrarily designated as U1, U2 …………………U7. These RNAs act as enzymes and participate in processing of mRNAs and are similar to ribosomes.
iii. Telomerase RNA:
It is found in eukaryotic nucleus and adds telomeric DNA sequences onto the ends of linear chromosome.
iv. Guide RNA:
This small type of RNA is found in Trypanosoma, a parasite causing African sleeping sickness. Insertion/ deletion editing in trypanosomes is directed by “gRNA” (guide RNA) template, which are transcribed from the mitochondrial genome.
v. Antisense RNA:
It is involved in gene regulation. Sometimes it is possible for the DNA strand, other than the DNA strand from which sense RNA has been transcribed to be copied into RNA. The RNA produced by transcription of ‘wrong’ strand of DNA is called antisense RNA.
It’s binding to sense RNA may physically block ribosome binding or elongation and thus may inhibit translation. Binding of antisense RNA to sense RNA may also trigger the degradation of sense RNA and thus block the gene expression.
vi. Sno RNA:
This small RNA occurs in nucleolus.
Properties of RNA:
The physical properties of RNA are almost like those of DNA. However, RNAs can absorb UV light more strongly than DNA. It is negative to Fuelgen reaction. The presence of the 2′ OH, enable the RNA to be degraded with alkali. RNA can hybridize with single stranded DNA to form DNA: RNA duplex. RNA can also act as genetic material in some viruses.