In this article we will discuss about the Translation in E.Coli:- 1. Introduction to Translation 2. Events of Translation 3. Translation of mRNA in E.Coli 4. Elongation of Polypeptide Chain 5. Termination of Translation.
Contents:
- Introduction to Translation
- Events of Translation
- Translation of mRNA in E.Coli
- Elongation of Polypeptide Chain
- Termination of Translation
1. Introduction to Translation:
Translation of mRNA is the biological polymerization of amino acids into polypeptide chains. The central question in translation is how triplet ribonucleotides of mRNA direct specific amino acids into their correct position in the polypeptide? This question was answered when transfer RNA (tRNA) was discovered.
In association with a ribosome, mRNA presents a triplet codon that calls for a specific amino acid. A specific tRNA molecule contains within its nucleotide sequence three consecutive ribonucleotides complementary to the codon (of mRNA), called the anticodon, which can base-pair with the codon. Another region of this tRNA is covalently bonded to its corresponding amino acid.
ADVERTISEMENTS:
Inside the ribosome, hydrogen bonding of tRNAs to mRNA holds the amino acid in proximity so that a peptide bond can be formed. As this process occurs over and over again and as mRNA runs through the ribosome, amino acids are polymerized into a polypeptide.
So, the process of translation largely depends on ribosomes and tRNAs. Translation involves a long sequence of events and the concerted activities of several associated factors.
2. Events of Translation:
Before describing the actual process of translation, other important events are being discussed as follows:
Activation of Amino Acids:
ADVERTISEMENTS:
Within the cytosol, amino acids react with ATP and is converted to activated form, aminoacylzadenylic acid. A covalent linkage is formed between the 5′-phosphate group of ATP and the carboxyl end of the amino acid. This molecule remains associated with the synthetase enzyme and form a complex which then reacts with a specific tRNA molecule.
Charging the tRNA:
Prior to translation, the tRNA molecules must be chemically linked to their respective amino acids. This process is called charging. It occurs under the direction of the enzyme, aminoacyl- tRNA synthetizes. During charging, the amino acid is transferred from the large enzyme bound complex to respective tRNA and bonded covalently to the adenine residue at the 3′ end.
Aminoacyl tRNA synthetizes catalyze this step. These enzymes are highly specific because they recognize only one amino acid and only a subset of corresponding tRNAs called iso-accepting tRNAs. This recognition is very crucial if fidelity of translation is to be maintained.
ADVERTISEMENTS:
Since there are twenty different amino acids involved in the formation of proteins in biological systems, there must be at least twenty different tRNAs and as many different enzymes.
However, because of the ability of the third member of a triplet code to ‘wobble’, it is now thought that there are at least 32 different tRNAs. It is also believed that there are only 20 synthetases, one for each amino acid, regardless of the greater number of corresponding tRNAs.
Dissociation of Ribosome:
The ribosomes of prokaryotes are 70S particles consisting of a large 50S subunit and a small 30S subunit. Before binding with mRNA, ribosomes get dissociated into two subunits and re-associated following the mRNA binding and again get dissociated after the termination of translation. Dissociation of subunits occurs in presence of a dissociation protein, located on the small subunit that requires Mg++ to be activated.
3. Translation of mRNA in E.Coli:
Three basic stages of translation are initiation, elongation and termination.
Initiation:
Initiator Codon and Initiator tRNA:
In both prokaryotes and eukaryotes, translation usually begins at the AUG initiator codon in the mRNA. It specifies the amino acid, methionine. However, in prokaryotes, the initiator methionine is modified, called formyl methionine (f Met), in which formyl group has been added to methionine’s amino group.
The first tRNA that brings this amino acid is called tRNA-f-Met, which has the anticodon 5′-CAU-3′ to bind to the AUG start codon.
ADVERTISEMENTS:
The amino-acylation of tRNA-f-Met occurs as follows: First, methionyl-tRNA synthetase catalyzes the addition of methionine to the tRNA. Then an enzyme called transformylase catalyzes the addition of the formyl group to the methionine. The resulting molecule is designated f-Met-tRNA-f- Met (it means that the tRNA is specific for the attachment of f-Met and that, in fact, f-Met is attached to it).
Initiation Factors:
Three main initiation factors (proteins) were isolated from the 30S subunit of ribosomes in prokaryotes and were referred to as IF-1, IF-2 and IF-3.
Interference Factors:
In E. coli several interference factors or I factors are found which may alter the specificity of initiator factors and thus regulate the rate of translation.
Formation of Initiation Complex:
Initiation of translation in E. coli involves the formation of initiation complex that contains mRNA, small ribosome subunit (30S), tRNA- f-Met, N formyl methionine and three initiation factors. In prokaryotes, initiation of protein synthesis requires other information coded in the base sequence of an mRNA molecule upstream (to the 5′ end) of the initiation codon AUG.
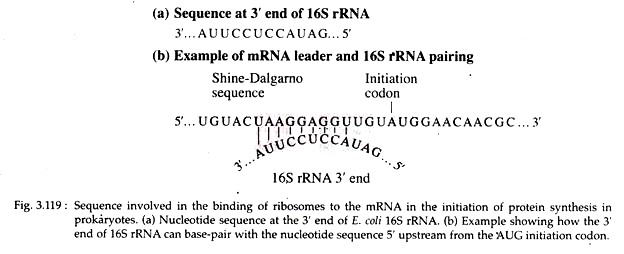
These sequences serve to align the ribosome on the message in the proper reading frame so that polypeptide synthesis can proceed correctly. Evidence from the work of John Shine and Lynn Dalgarno indicates that a purine rich sequence, 5′-UAAG- GAGGU-3′ (Shine-Dalgarno sequence) is present within the leader sequence of mRNA.
This sequence is complementary to a pyrimidine rich region, 3′-AUUCCUCCA-.5′ of 16S rRNA of 30S ribosomal subunit (Fig. 3.119).
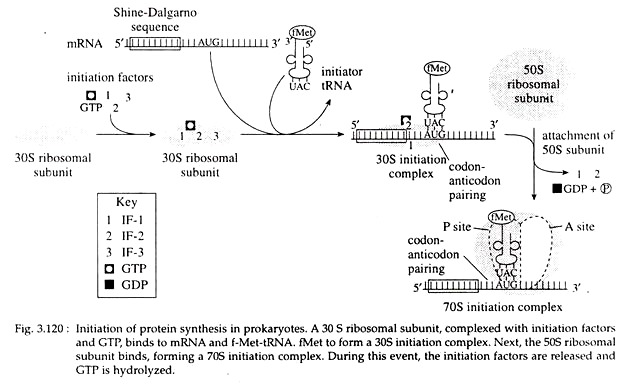
At the beginning of initiation, three initiation factors, IF-1, IF-2 and IF-3, are bound to the 30S ribosomal subunit along with a molecule of GTP. The f-Met-tRNA-f-Met and the mRNA then attaches to the 30S-IF-GTP complex to form the 30S initiation complex. IF-3 is then released as a result of this process.
Next, the 50S subunit binds, leading to GTP hydrolysis and the release of IF-1 and IF-2. The final complex is called the 70S initiation complex (Fig. 3.120). All three IF molecules are recycled for use in other initiation reactions. The 70S ribosome has two binding sites for aminoacyl-tRNA, the peptidyl (P), and aminoacyl (A) sites. The f-Met -tRNA-f Met is bound to the mRNA in the P site.
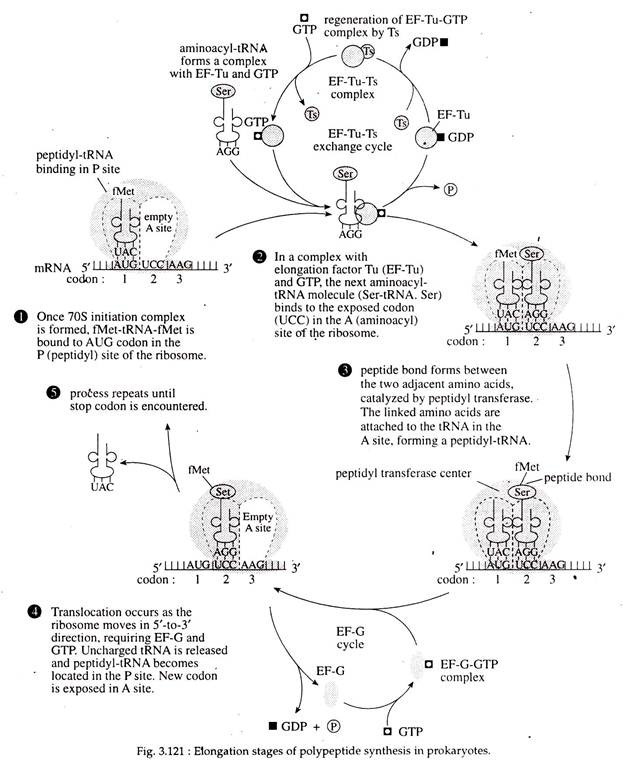
4. Elongation of Polypeptide Chain:
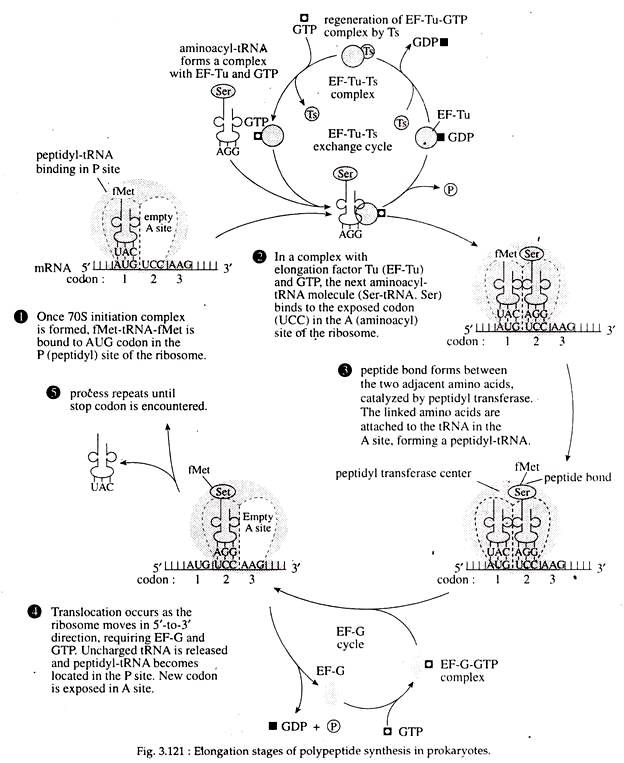
Binding of aminoacyl-tRNA (charged tRNA) to the ribosome: The new or second incoming aminoacyl-tRNA enters into the vacant A site of the ribosome and locates itself on the second codon of the mRNA with its antiocodon. The P site of the ribosome at this point is occupied by Met-tRNA.
The entry of aminoacyl tRNA into A site of ribosome is mediated by the cytoplasmic elongation factor IF-T, which has two sub- units EF-Tu (temperature unstable) and EF-Ts (temperature stable). At first EF-Ts, binds to EF-Tu and displaces the GDP. Next, GTP binds to the EF-Tu-EF-Ts complex to produce an EF-Tu-GTP complex, simultaneously with the release of EF-Ts.
The amino- acyl-tRNA binds to the EF-Tu-GTP, and that complex can then bind to the A site in the ribosome when the appropriate codon is exposed. When the aminoacyl-tRNA binds to the codon in the A site, the GTP is hydro- lyzed into GDP and Pi. The released EF-Ts then becomes involved in regeneration of EF-Tu (Fig. 3.121).
Peptide Bond Formation:
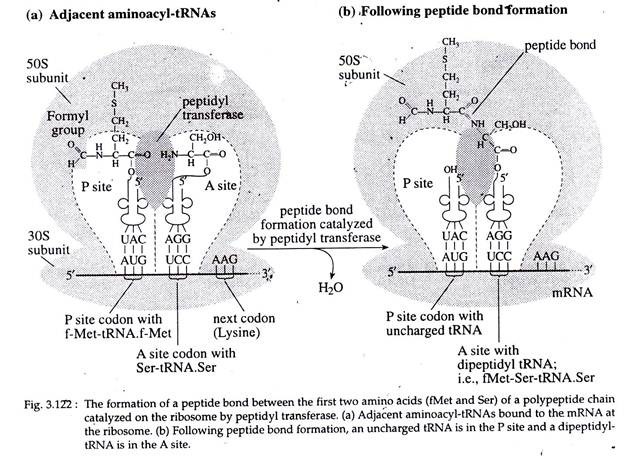
The next step involves formation of the first peptide bond between Met of the initiating tRNA and the amino acid of the second aminoacyl-tRNA (in this case Ser-tRNA. ser). First, a breakage the f-Met and its tRNA.
Next, a peptide bond of the bond between the carboxyl group of is formed between the now-freed f-Met and the amino acid and the tRNA in the P site the ser (in this case) attached to the tRNA in occurs. In this case, the breakage is between the A site. This reaction is catalyzed by the enzyme, peptidyl transferase, a part of 23S rRNA located at 50S subunit of ribosome (Fig. 3.122).
Once the peptide bond has formed (Fig. 3.121) the uncharged tRNA (tRNA without an attached amino acid) is left in the P site. The t-RNA in the A-site, now called peptidyl- t-RNA, has the first two amino acids of the polypeptide chain attached to it (in this case f-Met-Ser) (Fig. 3.122).
Translocation:
Now the whole ribosome moves along the mRNA in 5′ —> 3′ direction of the mRNA for a distance of another codon or base triplet. This is called translocation. As a result, the di-peptidyl-tRNA comes to the P site and the empty tRNA is dislodged from P site and released.
The A site is occupied by a third codon of mRNA. Such translocation requires specific elongation factor EF-G (a protein of 72,000 Dalton) that binds to ribosome and mediates its movement along mRNA. This is accompanied by hydrolysis of GTP (Fig. 3.121, 122).
Continuation of Chain Elongation:
Now the aminoacyl-tRNA that entered the A-site gets bind to the next codon of mRNA. The dipeptide residue from the di-peptidyl-tRNA at P site is peptide bonded to the amino acid brought in by the next tRNA.
Again translocation occurs and the empty tRNA gets released from the P site. In this way, chain elongation goes on. A tunnel exists within the large subunit of the ribosome through which the elongated polypeptide emerges. In E. coli, elongation occurs at a rate of about 15 amino acids per second at 37°C.
Once the ribosome moves away from the initiation site on mRNA, the initiation site is open for another initiation event to occur. Thus, many ribosomes may simultaneously translate a single mRNA. This complex of mRNA molecule and all the ribosomes is called a polyribosome or polysome.
5. Termination of Translation:
The termination of translation is signaled by one or more of three triplet codes present in the A-site. These are UAG (amber), UAA (ochre) or UGA (Opal). These codons do not specify any amino acid and thus do not call for a tRNA in the A-site. These codons are called stop codons or termination codons or nonsense codons.
The ribosome recognizes a termination codon only with the help of some proteins called termination factors or release factors (RF), which read the chain termination codons (Fig. 3.123b). Three RFs are RF-1, RF-2 and RF-3 of which RF-1 recognizes UAA and UAG, RF-2 recognizes UAA and UGA, and factor RF-3 does not recognize any codon but stimulates the termination events.
During termination, the GTP dependent release factors cleave the polypeptide from the terminal tRNA in the P site of the ribosome and then releases it from the translation complex. Following this cleavage, tRNA is released from the ribosome which then dissociates into its subunits (Fig. 3.123). The initiating amino acid f-Met is cleaved from the completed polypeptide.
When any termination codon appears in the middle of an mRNA due to its mutation, the polypeptide chain is prematurely terminated. Again if there is a UGA codon in an appropriate nucleotide context located internally in the mRNA, the modified amino acid selenocystein can be incorporated into the protein with the help of a special tRNA having the anticodon 5′-UCA-3′.
In sum, protein synthesis is a complex process involving interaction between three major classes of RNA (mRNA, rRNA and tRNA) and a large number of accessory protein factors that catalyze the process. By repeated translation of an mRNA molecule with a polysome, a large number of identical protein molecules can be produced.
Thus, from a single gene, large quantities of a protein can be produced by two amplification steps:
(i) The production of multiple mRNAs from the gene and
(ii) The production of many protein molecules by repeated translation of each mRNA.